One of my first tasks as a new post-doc was to undertake a systematic quantitative literature review. We wanted to get a feel for the international & NZ literature on functional biodiversity in agroecosystems, and this was a bit daunting for me as I my background is in invasions, not native biodiversity or agriculture! Luckily, the review method we chose relies on data, not expert knowledge - we chose the method developed by Griffith University (https://www.griffith.edu.au/griffith-sciences/school-environment-science/research/systematic-quantitative-literature-review).
It's quite an exhaustive process but the method does a really good job of catching easily overlooked papers, and provides a reproducible and transparent method for conducting reviews and meta-analyses. I'm a fan!
The first few steps involve defining your keywords and databases for undertaking the searches, and designing a way of storing your papers and extracting the data. Once you've completed the first 10% of your search, though, you'll need to do a stock-take of the papers that you're picking up and make sure you haven't missed any key words in your search terms.
It was at this point that I realized two things: a) automating this would save me a lot of time, and b) there were no existing programs that could do what I wanted. So, I wrote up some python code to read in all papers, extract the keywords and write them to a new text file. I then used R to rank them by how commonly they occurred and graph the results, and added any commonly-occurring keywords to my database search strings.
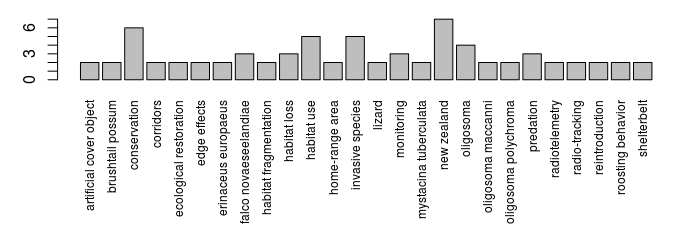
It's not the prettiest graph I've ever made! These were all keywords that occurred more than once in my database. We added "corridor" and "habitat" to our searches based on this.
You can download a copy of my code at my GitHub https://github.com/pannellj/systematicreviews (but disclaimer! I am a total python newbie and it's a bit rough. Suggestions, commits are very welcome). To run the code, you'll first need to convert the PDFs of your papers to text files. I did this using the command line utility pdftotext. The bash script is also available on GitHub.
My next post will be about the next step in the literature review, which was a bit trickier - cross-checking the papers in the database using python. Let me know in the comments if you've found this useful!

Write a comment
ISABEL CRUZ (Friday, 08 January 2021 07:28)
Hi!
Thanks for sharing your experience. I am learning Python to use it for Nursing Evidenced Based studies.
I will try your code.
regards!